Running AutoDock Vina via MCP
Model Context Protocol (MCP)
Reference 1: zenn
Reference 2: DeepLearning.ai
When an MCP client (like Claude Desktop or Cursor) sends a request to an MCP server, the server accesses its stored information and returns search results. The advantage is that LLMs can access external information.
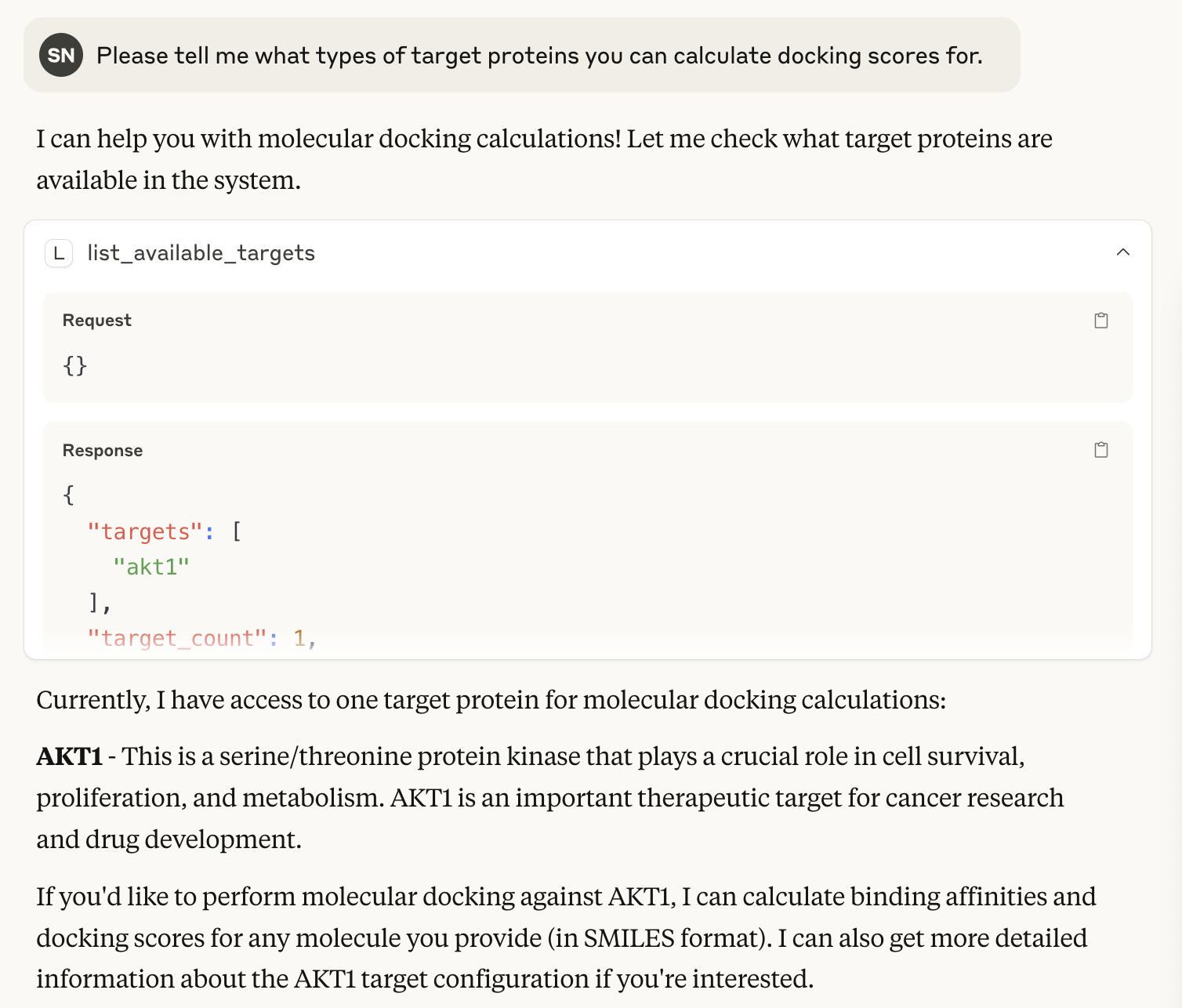
Docking calculations from Claude Desktop using SMILES and target names
To get hands-on experience, I had docking scores calculated using Vina through MCP. The MCP server resources only contain target protein names and config information (pocket center, grid size, etc.), and when I specify SMILES and predefined target names in Claude Desktop, docking calculations run automatically.
Source code:
https://github.com/shogo-d-nakamura/MCP_Vina
References:
Claude document
chatMol
Blog
Downloading Claude Desktop
Claude provides documentation on how to set up MCP servers (link above). Following this, I set up Claude Desktop. The config JSON can be ported to work with Cursor and other clients as well.
Creating a virtual environment with uv
1
curl -LsSf https://astral.sh/uv/install.sh | sh
After curl, restart the terminal to enable uv.
Since the published repository contains pyproject.toml:
1
uv pip install -e .
This installs all packages.
To manage packages from scratch, use uv init followed by uv add for necessary additions, as mentioned in Claude’s documentation. You can specify Python version with uv init $DIR_NAME -p 3.10.
1
2
3
4
5
uv init test -p 3.10
cd test
source .venv/bin/activate
# install packages
uv add httpx rdkit meeko "mcp[cli]"
I used scrubber (now molscrub) to generate 3D structures from SMILES. If scrubber’s scrub.py and meeko’s mk_prepare_ligand.py are in PATH, ligand preprocessing can be performed.
1
2
git clone --single-branch --branch develop https://github.com/forlilab/scrubber.git
cd scrubber; uv pip install -e .; cd ..
Claude Desktop Settings
In Claude Desktop Settings -> Developer -> Edit Config, a JSON file opens. Copy and paste the following: 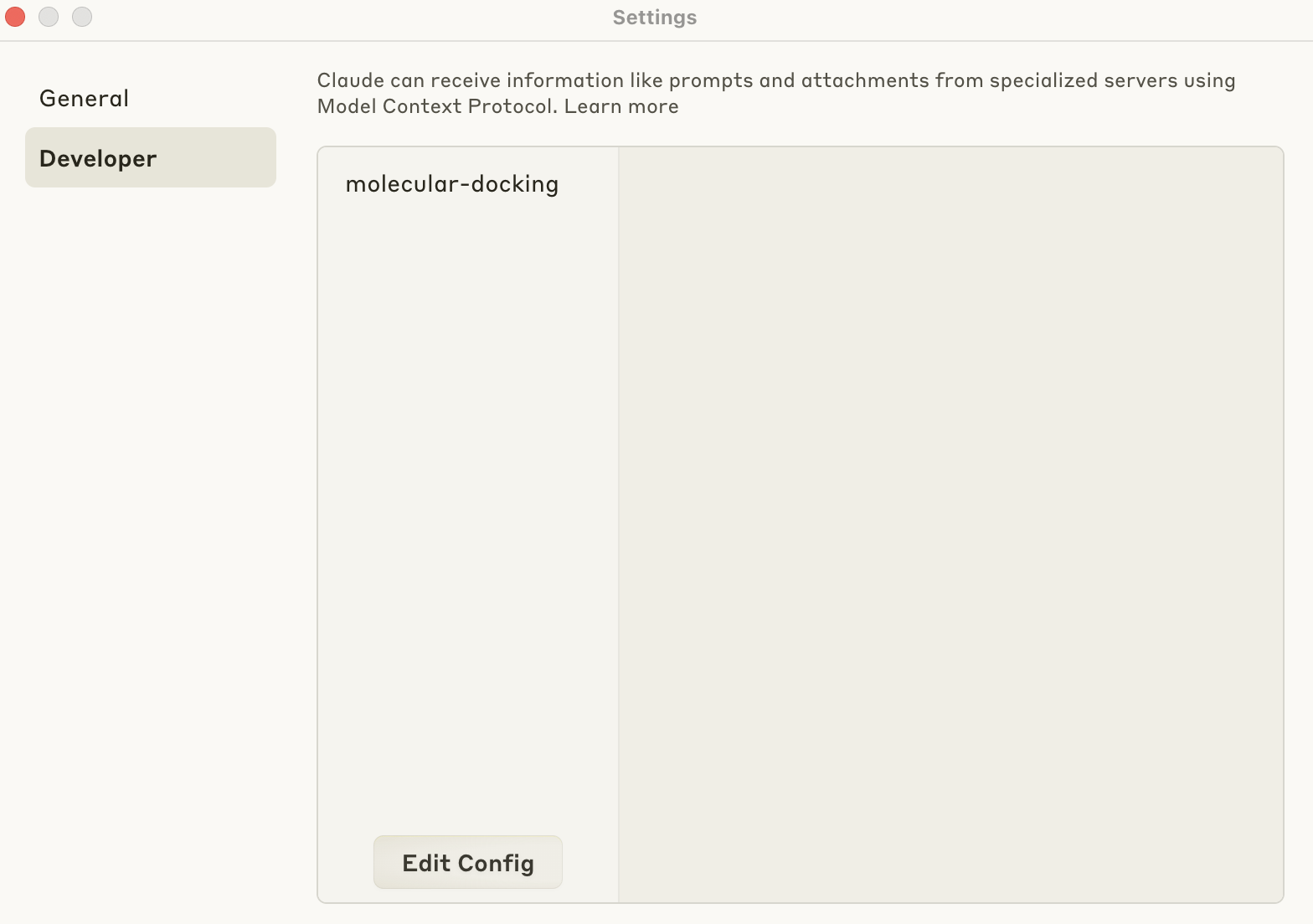
1
2
3
4
5
6
7
8
9
10
11
12
13
14
{
"mcpServers": {
"molecular-docking": {
"command": "uv",
"args": [
"--directory",
"/PATH/TO/molDocking",
"run",
"server.py"
]
}
},
"globalShortcut": ""
}
After updating Settings, restart Claude.
Claude’s documentation suggests this should work, but I encountered errors (MacBook M4, Sequoia). It seemed uv wasn’t recognized, so entering the absolute path obtained with which uv made it work properly. This JSON can be copied to other MCP client configs for the same connection. Standardization is awesome!
1
2
3
4
5
6
7
8
9
10
11
12
13
14
{
"mcpServers": {
"molecular-docking": {
"command": "/PATH/TO/uv",
"args": [
"--directory",
"/PATH/TO/molDocking",
"run",
"server.py"
]
}
},
"globalShortcut": ""
}
Usage
Target protein types
When asked about registered target proteins, AKT1 was returned. This runs the list_available_targets function with the @mcp.tool() decorator in /molDocking/server.py. Since only AKT1’s pdbqt and config information are pre-stored, it returns that.
1
2
3
@mcp.tool()
def list_available_targets() -> Dict[str, Any]:
~
config.txt information
Functions that output pocket center coordinates are also written with the @mcp.tool() decorator. 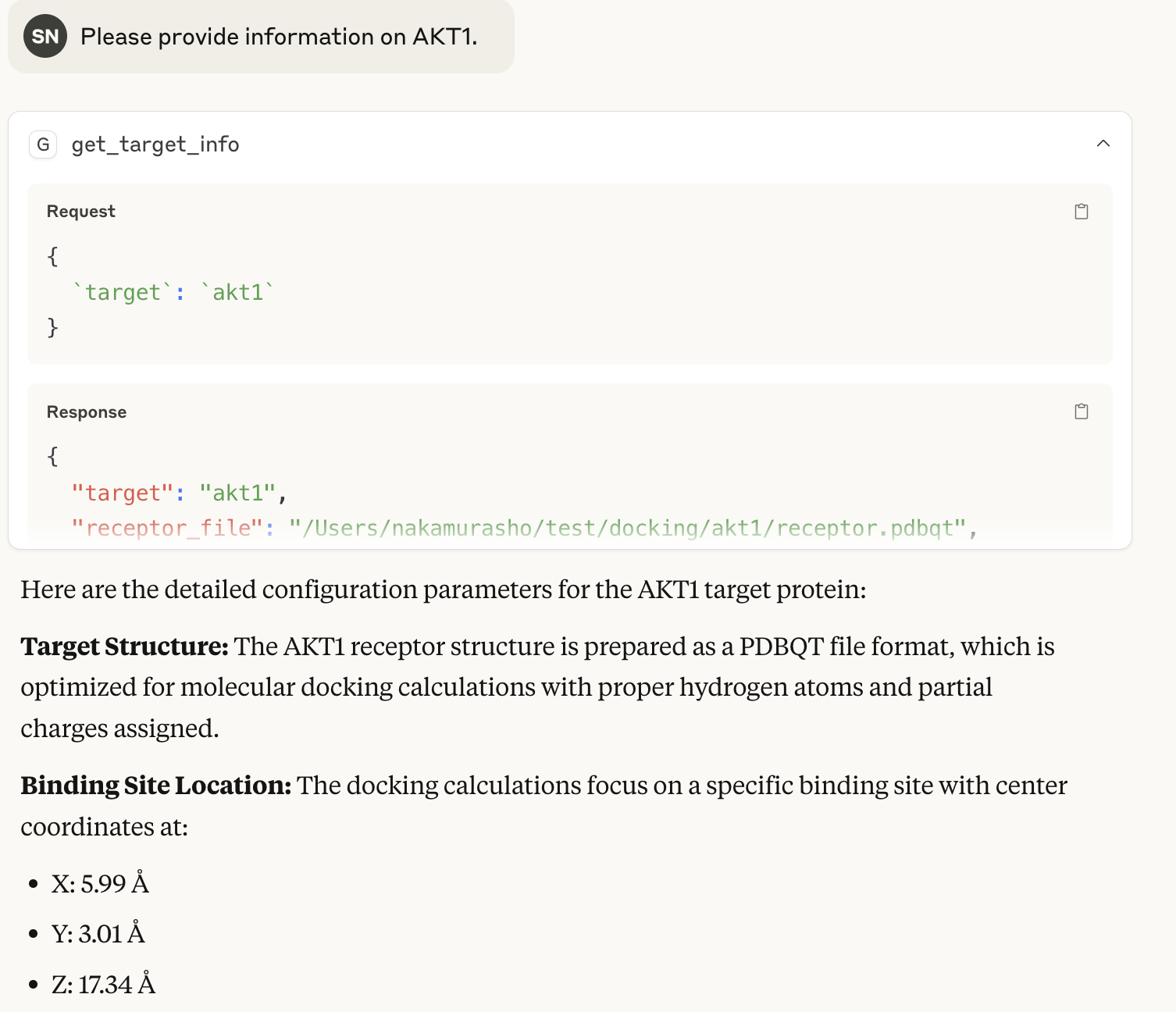
Running actual calculations
I had random SMILES docked to AKT1. The @mcp.tool() run_molecular_docking() is executed. 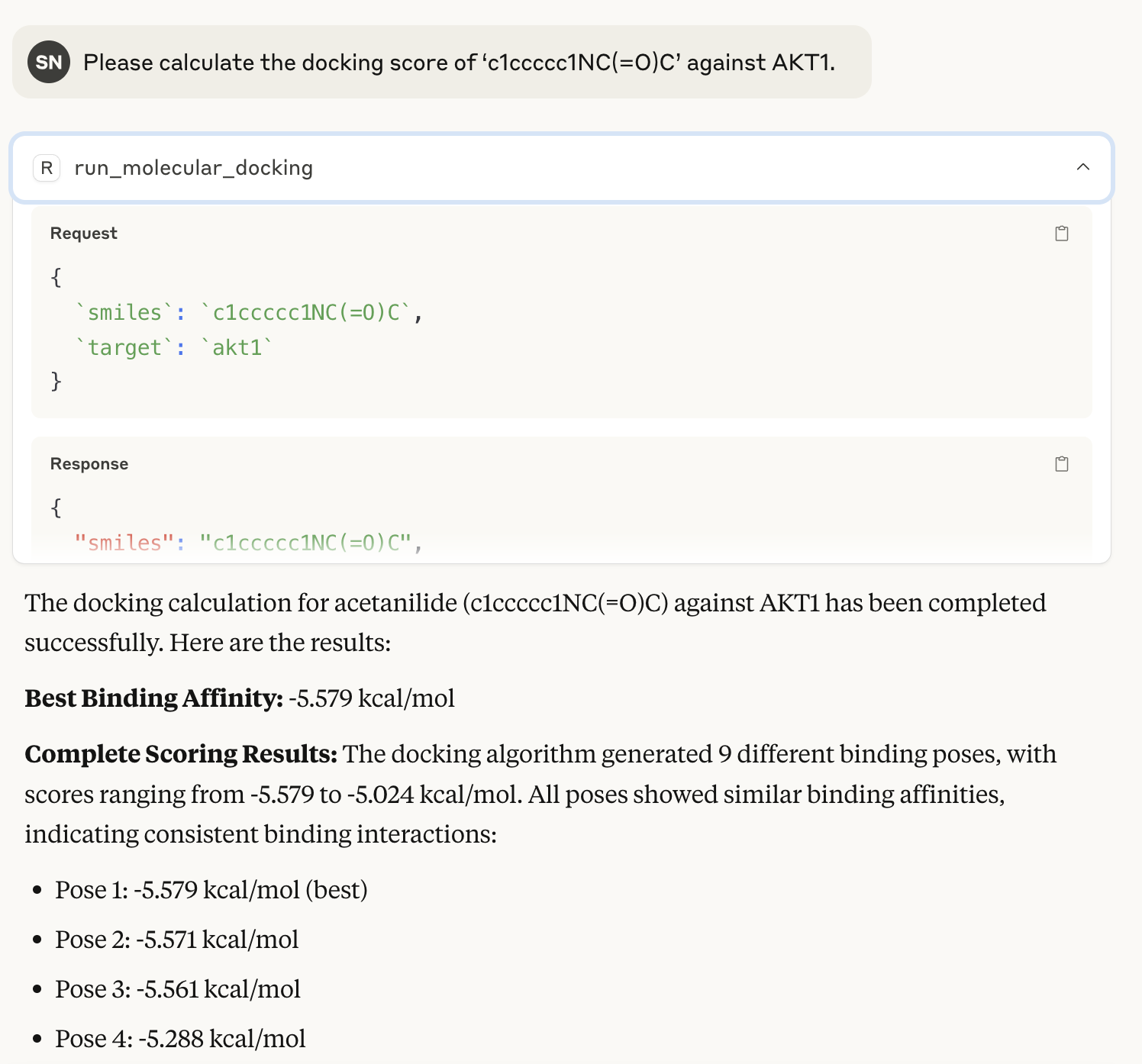 Though not particularly useful, Claude provides its own interpretation as well.
Though not particularly useful, Claude provides its own interpretation as well. 
This setup now returns Vina scores when target protein names and SMILES registered beforehand are provided. While this time I only placed protein pdbqt and config in MCP, combining local files and REST APIs could enable molecular design-like tasks using LLM as a wrapper.